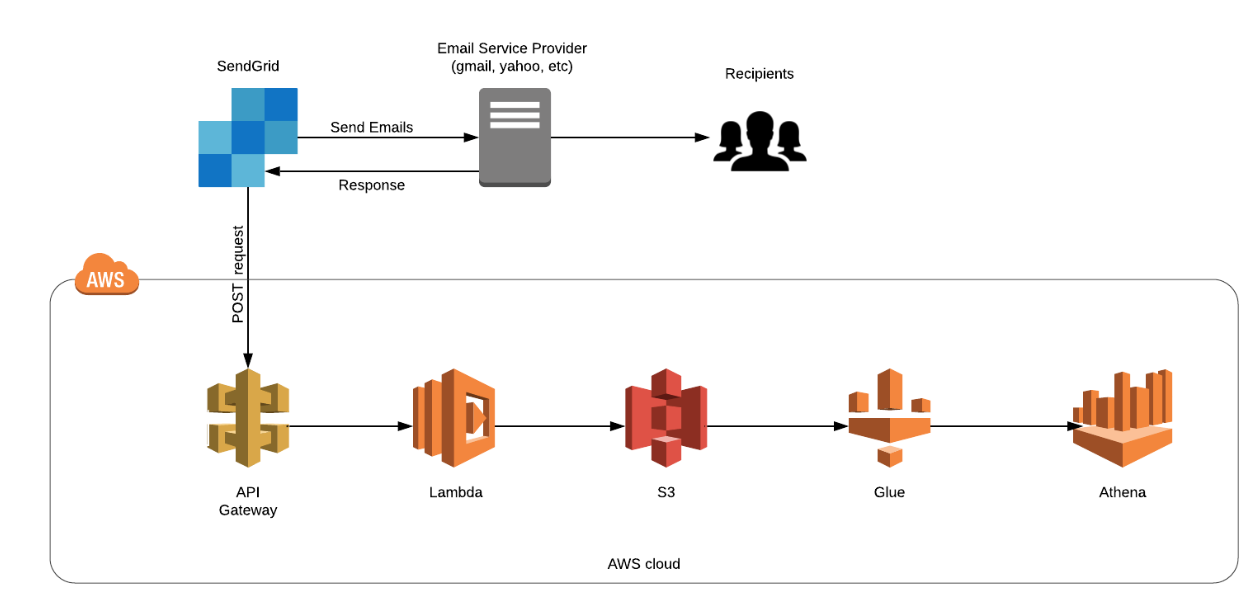
Understanding Partition Projections in AWS Athena
If you are somebody who uses AWS Athena to query large highly partitioned tables on a daily basis you must know how difficult it is to maintain the partitions. As your partitions grow, you also need to update the metadata in Glue Data Catalog, or else the new data isn’t scanned. Some of us even... » read more